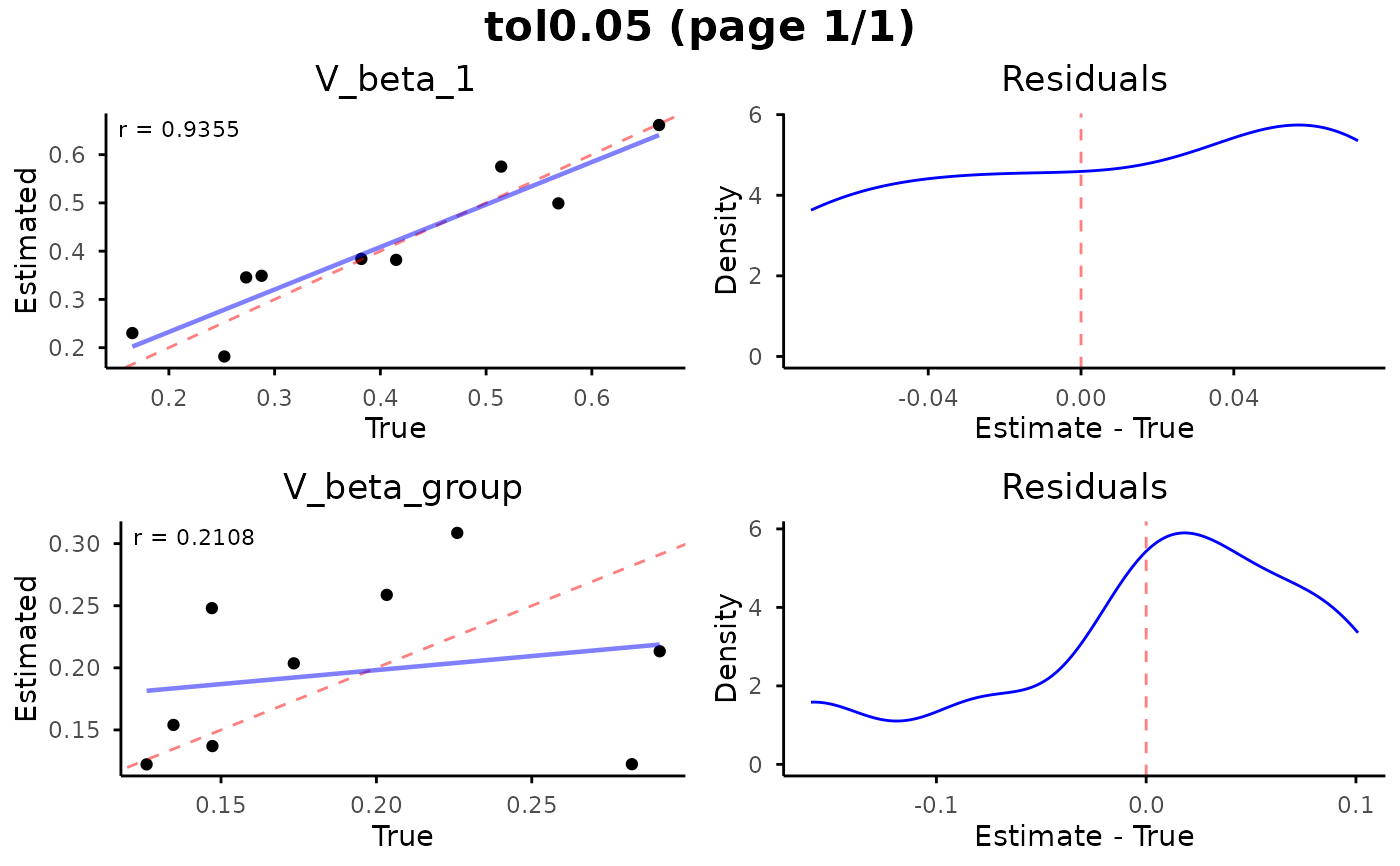
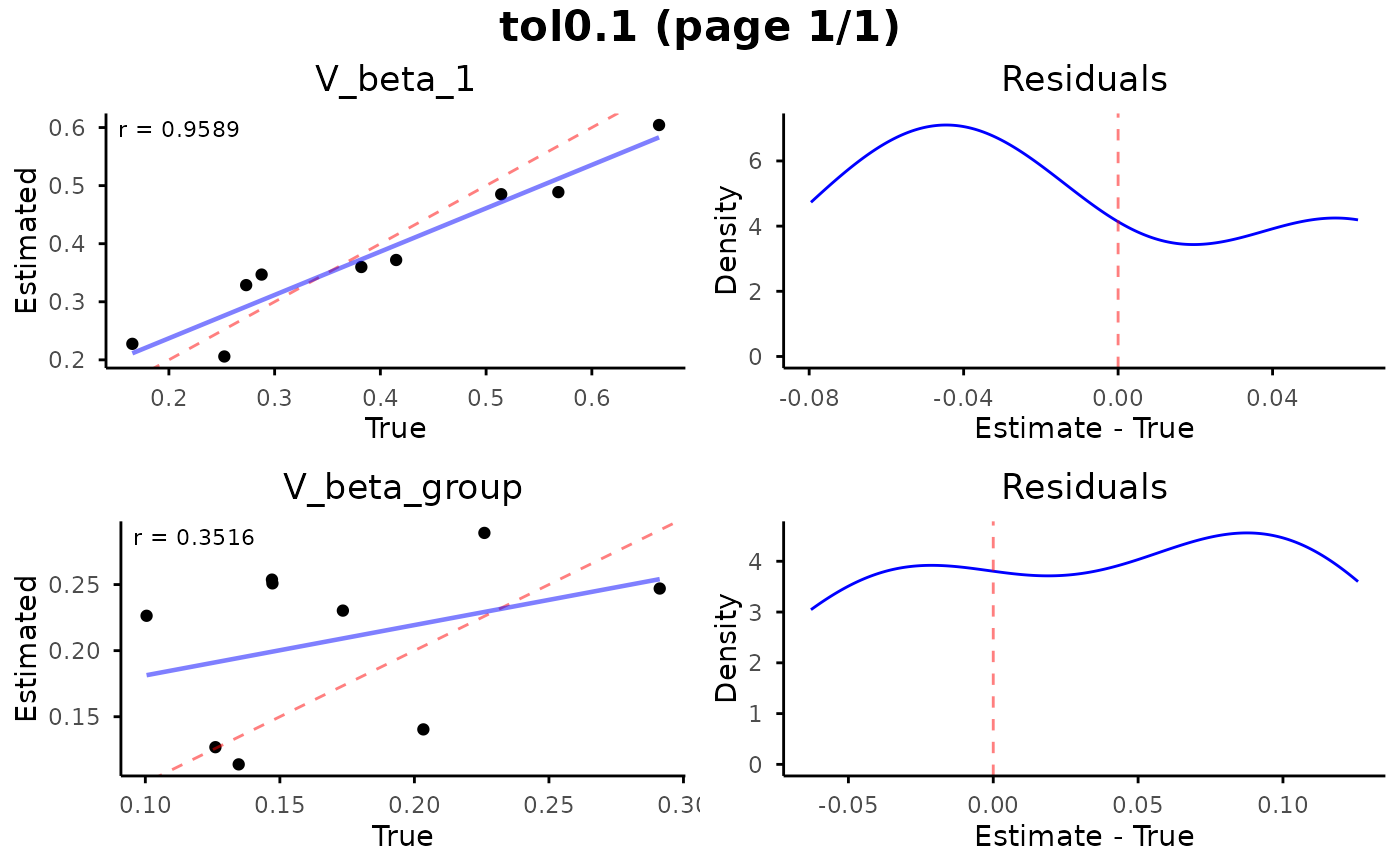
Visualize parameter recovery from cross-validation results, showing estimated vs. true parameter values and residual distributions for each parameter.
Arguments
- data
A
cv4abcobject containing true parameters and cross-validated estimates.- ...
Additional arguments:
- n_rows
Integer; number of rows in the plot grid (default: 3)
- n_cols
Integer; number of columns in the plot grid, multiplied by 2 for paired plots (default: 1)
- method
Character; smoothing method for
geom_smooth(default: "lm")- formula
Formula; used in
geom_smooth(default: y ~ x)- resid_tol
Numeric; quantile threshold for filtering residuals by absolute value. If specified, only observations with residuals below this quantile are plotted (default: NULL, no filtering)
- interactive
Logical; whether to pause between pages and wait for user input (default: FALSE)
Examples
# Load CV output from saved file
cv_file <- system.file(
"extdata", "rdm_minimal", "abc", "cv", "neuralnet.rds",
package = "eam"
)
abc_neuralnet_cv <- readRDS(cv_file)
# Plot parameter recovery
plot_cv_recovery(
abc_neuralnet_cv,
n_rows = 2,
n_cols = 1,
resid_tol = 0.99
)